TCGA is a valuable public database for cancer and pharmaceutical research. If you are new to this public dataset, it may not be obvious to you how to start downloading the data.
In this tutorial, you will learn how to download TCGA data.
What is TCGA?
Before downloading the TCGA data, you must know what is available within the TCGA dataset and whether you have the permission to download it.
The Cancer Genome Atlas (TCGA) is a public cancer database spearheaded by the National Cancer Institute and the National Human Genome Research Institute.
Started in 2006, TCGA has profiled over 11,000 tumors and normal biopsy samples across more than 30 cancer types.
Data types
The data covers a wide range of modalities, including genomic (DNA), epigenomic (methylation), transcriptomic (RNA), and proteomic data.
The TCGA effort harmonizes data generation, analysis pipelines, and clinical annotations. It is a valuable and free resource for researchers to uncover molecular drivers of malignancy, identify potential biomarkers, and guide the development of precision oncology strategies. Access to raw and processed data is provided through portals such as the Genomic Data Commons, facilitating reproducible, large-scale cancer research.
Cancer types
Multiple projects generate the TCGA data, and most of these projects study cancer samples originating from a single tissue type. For example, TCGA-BRCA studies breast cancer samples and compares them with normal breast tissues from the same patient. (As you may wonder, this can create questions about whether they are truly normal samples. But I will save this topic for another post.
Below is the full list of TACG projects/cancer types.
| Project Code | Cancer Type |
|---|---|
| TCGA-ACC | Adrenocortical Carcinoma |
| TCGA-BLCA | Bladder Urothelial Carcinoma |
| TCGA-BRCA | Breast Invasive Carcinoma |
| TCGA-CESC | Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma |
| TCGA-CHOL | Cholangiocarcinoma |
| TCGA-COAD | Colon Adenocarcinoma |
| TCGA-DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma |
| TCGA-ESCA | Esophageal Carcinoma |
| TCGA-GBM | Glioblastoma Multiforme |
| TCGA-HNSC | Head and Neck Squamous Cell Carcinoma |
| TCGA-KICH | Kidney Chromophobe |
| TCGA-KIRC | Kidney Renal Clear Cell Carcinoma |
| TCGA-KIRP | Kidney Renal Papillary Cell Carcinoma |
| TCGA-LAML | Acute Myeloid Leukemia |
| TCGA-LGG | Brain Lower Grade Glioma |
| TCGA-LIHC | Liver Hepatocellular Carcinoma |
| TCGA-LUAD | Lung Adenocarcinoma |
| TCGA-LUSC | Lung Squamous Cell Carcinoma |
| TCGA-MESO | Mesothelioma |
| TCGA-OV | Ovarian Serous Cystadenocarcinoma |
| TCGA-PAAD | Pancreatic Adenocarcinoma |
| TCGA-PCPG | Pheochromocytoma and Paraganglioma |
| TCGA-PRAD | Prostate Adenocarcinoma |
| TCGA-READ | Rectum Adenocarcinoma |
| TCGA-SARC | Sarcoma |
| TCGA-SKCM | Skin Cutaneous Melanoma |
| TCGA-STAD | Stomach Adenocarcinoma |
| TCGA-TGCT | Testicular Germ Cell Tumors |
| TCGA-THCA | Thyroid Carcinoma |
| TCGA-THYM | Thymoma |
| TCGA-UCEC | Uterine Corpus Endometrial Carcinoma |
| TCGA-UCS | Uterine Carcinosarcoma |
| TCGA-UVM | Uveal Melanoma |

TCGA Data level
TCGA data is available in 4 levels of processing:
- Level 1 – Raw Data: Unprocessed data straight from the instrument. E.g., FASTQ files from DNA sequencers. They can contain technical biases that are specific to the instrument platforms.
- Level 2 – Normalized Data: Platform-specific technical bias removed. E.g., Gene expression intensities normalized to control probes.
- Level 3 – Aggregated Data: Data summarized in a matrix format across samples and features. E.g., A two-dimensional matrix with columns representing patient samples and rows representing beta values from methylation probes.
- Level 4 – Regions of interest or derived calls: Analytical result derived from lower-level data. E.g., A list of mutated genes for a particular cancer type.
Level 1 data typically requires authorized (controlled) access because it can potentially be used to identify the patient.
Downloading TCGA data
The easiest way to start with TCGA analysis is by using the level 3 data. You will work with normalized, aggregated, and open data so that you can dive right into your research questions.
While all data can be found on the GDC Data Portal, it is not the easiest interface to navigate. I will describe two ways to download TCGA level 3 data
- XenaBrowser: Easy to use. Download data in a fixed matrix format through a web interface.
- TCGAbiolinks: An R library for building a query and downloading TCGA data. It’s a bit of work, but you can customize what to download.
XenaBrowser
The XenaBrowser, hosted by UC Santa Cruz, is one of the easiest ways to download level 3 TCGA data.
- Visit the Xena dataset page.
- Select the project you are interested in. For example, TCGA Pan-cancer.
- You will see the Pan-cancer datasets organized by data type, e.g., copy number, DNA methylation, etc. Download the dataset you are interested in.
- The patient metadata is under phenotype.
TCGAbiolinks (R)
The TCGAbiolinks library in R offers a programmatic interface to download the TCGA data. It’s a bit of work, but unlike XenaBrowser, you can pick and choose which patient sample to download.
1. Install TCGAbiolinks in R.
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("TCGAbiolinks")2. Check available projects.
projects <- TCGAbiolinks::getGDCprojects()
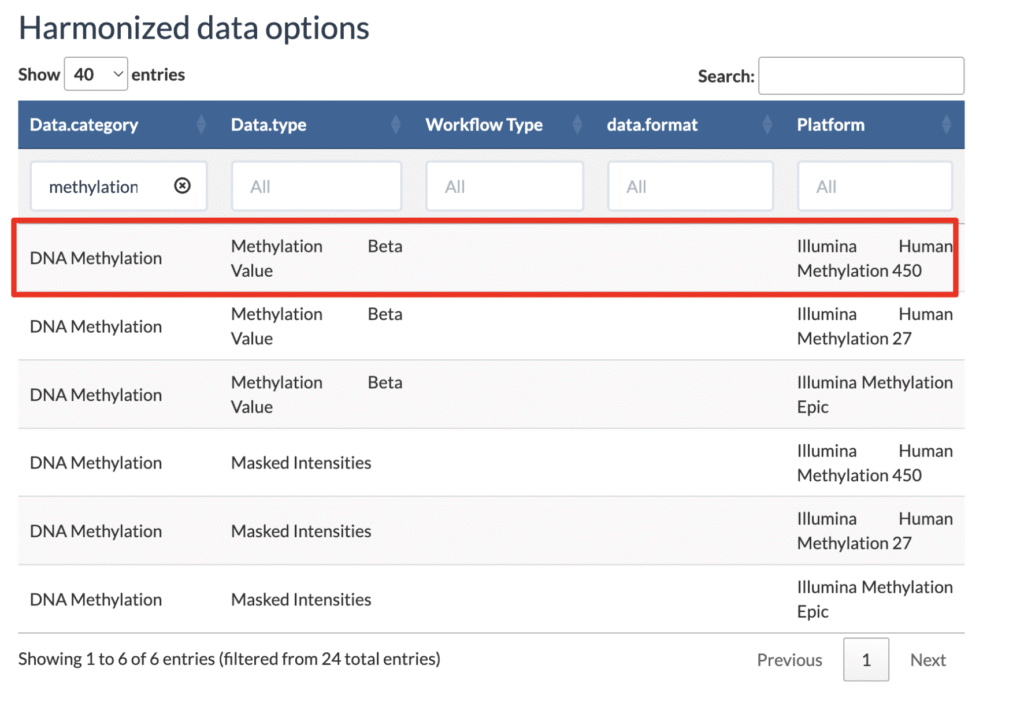
head(projects$project_id)3. Determine the data category and data type using the harmonization data option table. For example, after filtering the data category with the word “methylation”, you can see the following valid combination:
- data.category: DNA Methylation
- data.type: Methylation Beta Value
- platform: Illumina Human Methylation 450
You will need to know that the beta value is a normalized data type (level 2 or 3).
4. Build the query. See the documentation for argument options. The following command won’t download the data yet.
query <- GDCquery(
project = "TCGA-BRCA", # Replace with your project of interest
data.category = "DNA Methylation",
data.type = 'Methylation Beta Value',
platform = 'Illumina Human Methylation 450'
)5. Check the metadata of the query to make sure this is what you want to download.
getResults(query)[1:5,]6. Download the TCGA data.
GDCdownload(query)
data <- GDCprepare(query)Reference
- The Cancer Genome Atlas Pan-Cancer analysis project | Nature Genetics (2013) – This article summarizes the TCGA Pan-Cancer project.
- GDC Data Portal – The central repository of TCGA data.
